About
The GEM × Adaptyv RBX-1 Binder Design Competition 2026 brings together the computational protein design community to tackle a challenging and biologically significant target. Submit your designs—we'll test a total of 300 binders in the wet lab and announce results at ICLR 2026.
Why RBX-1?
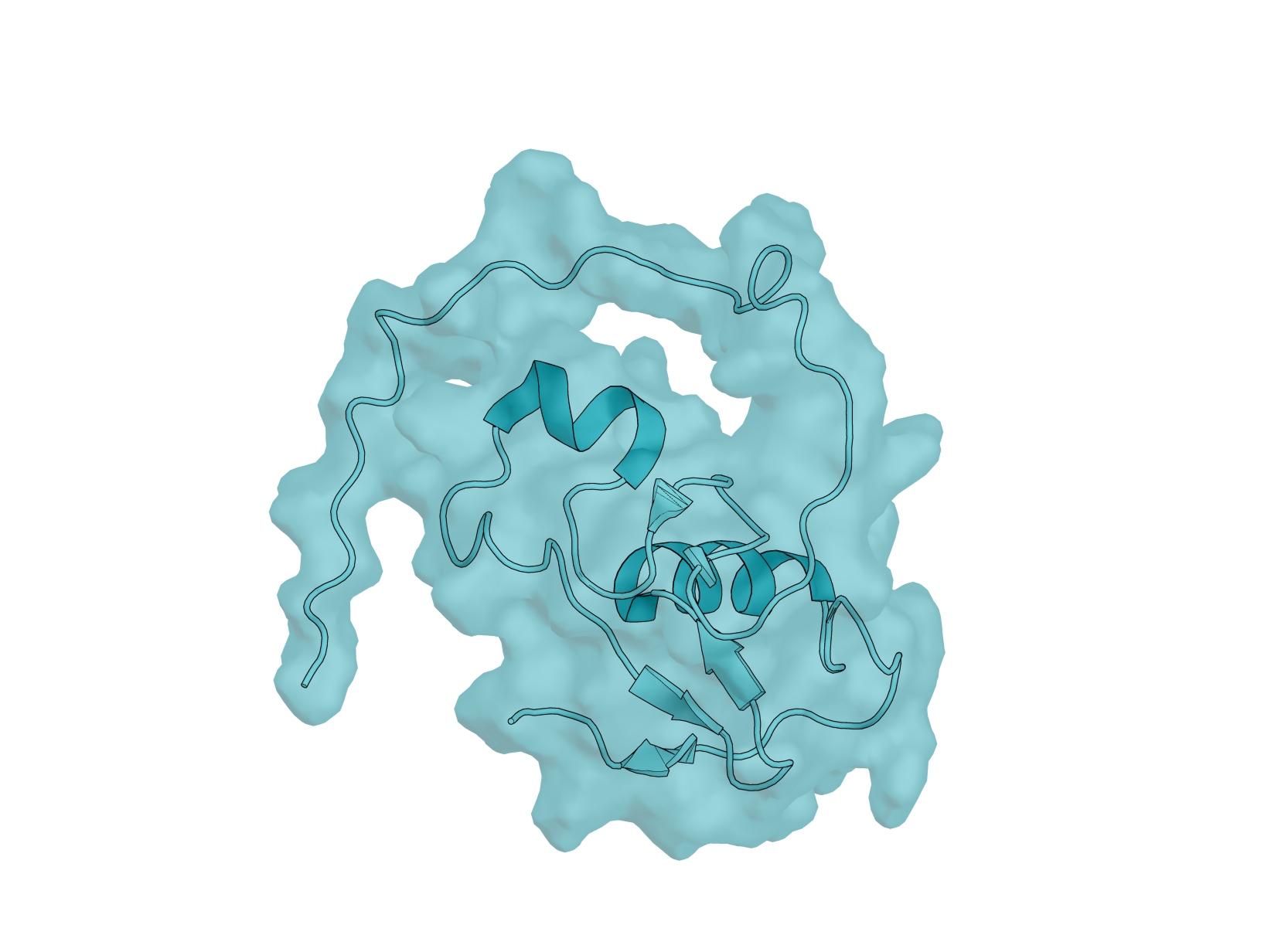
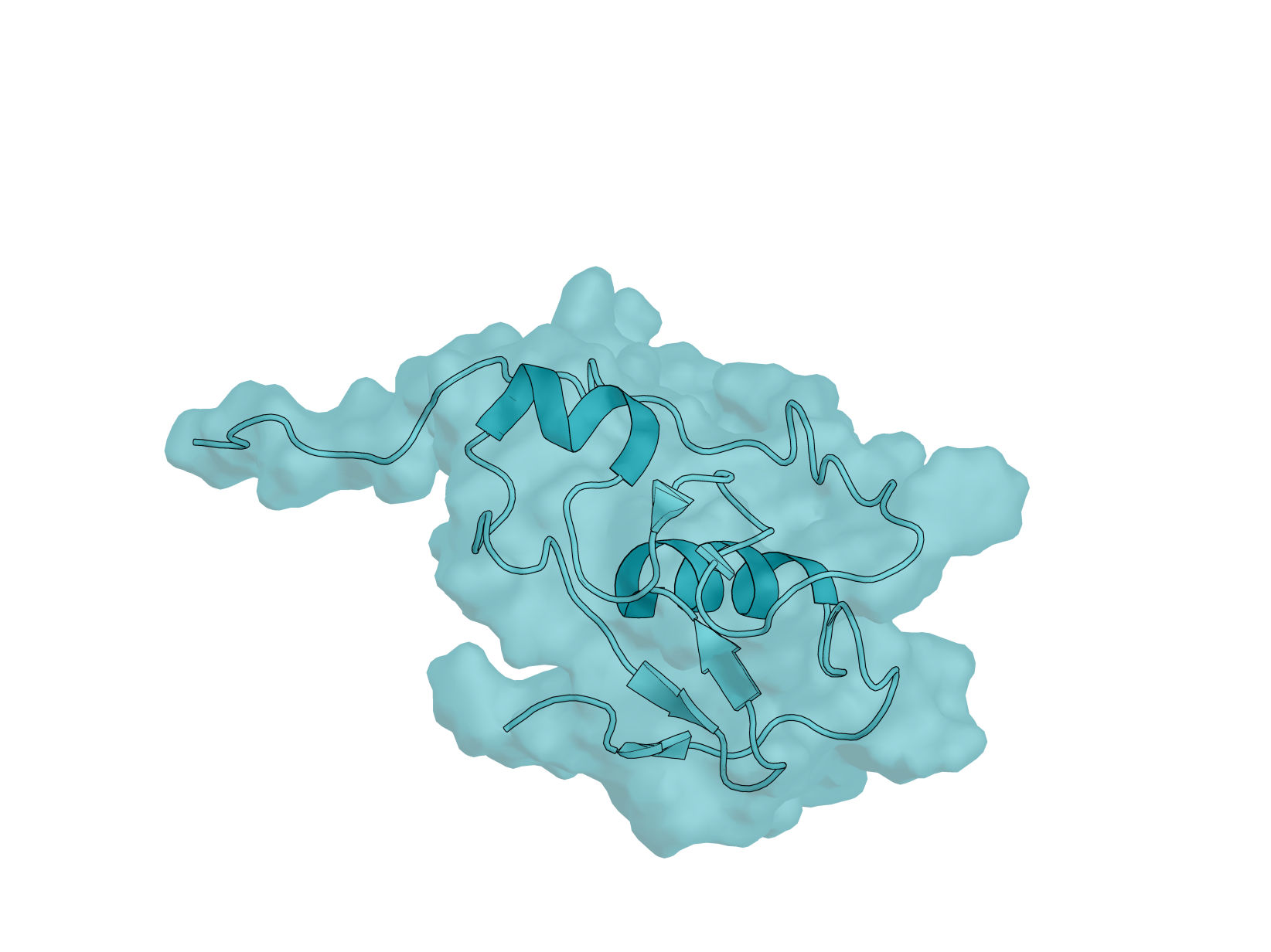
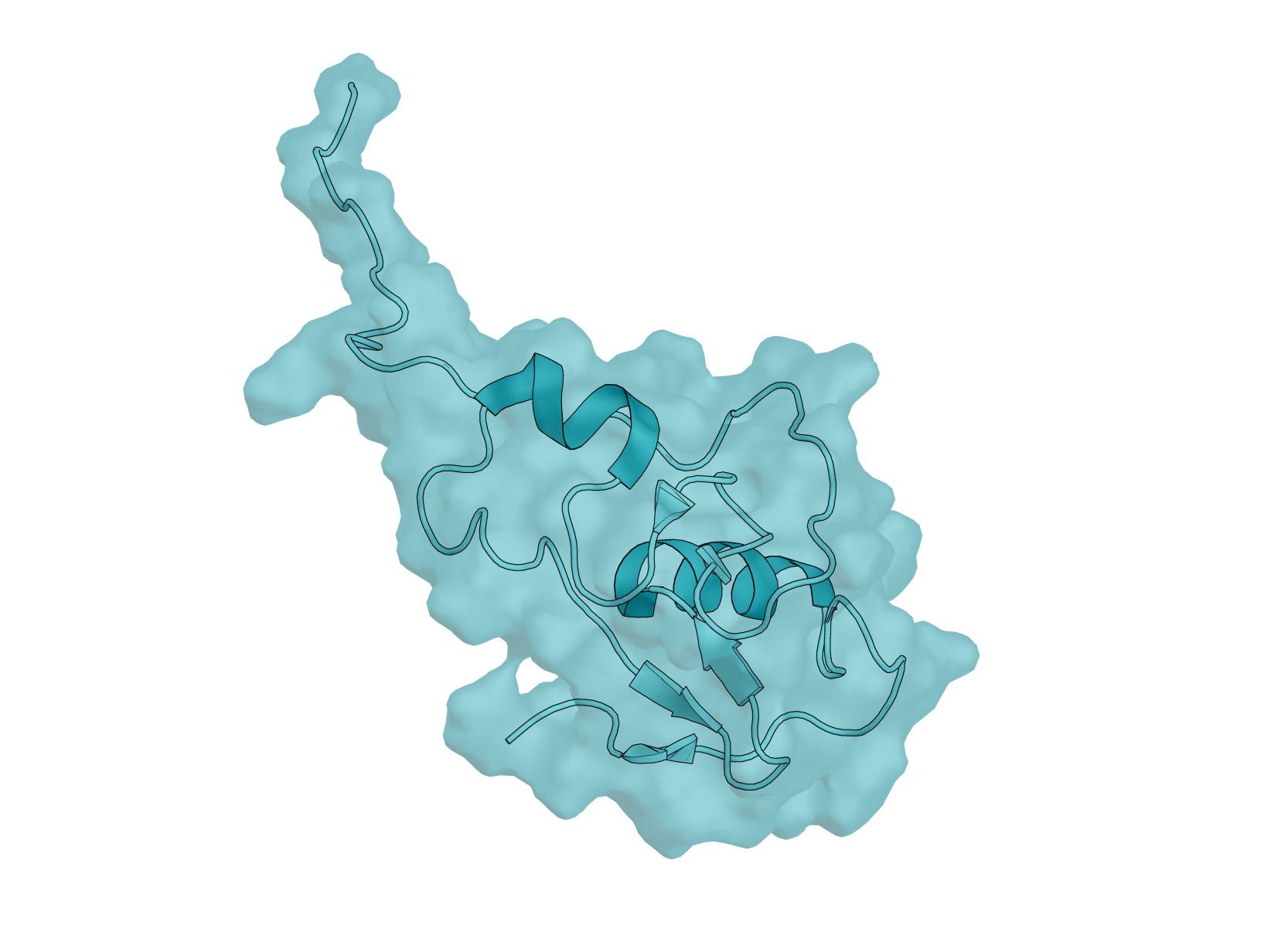
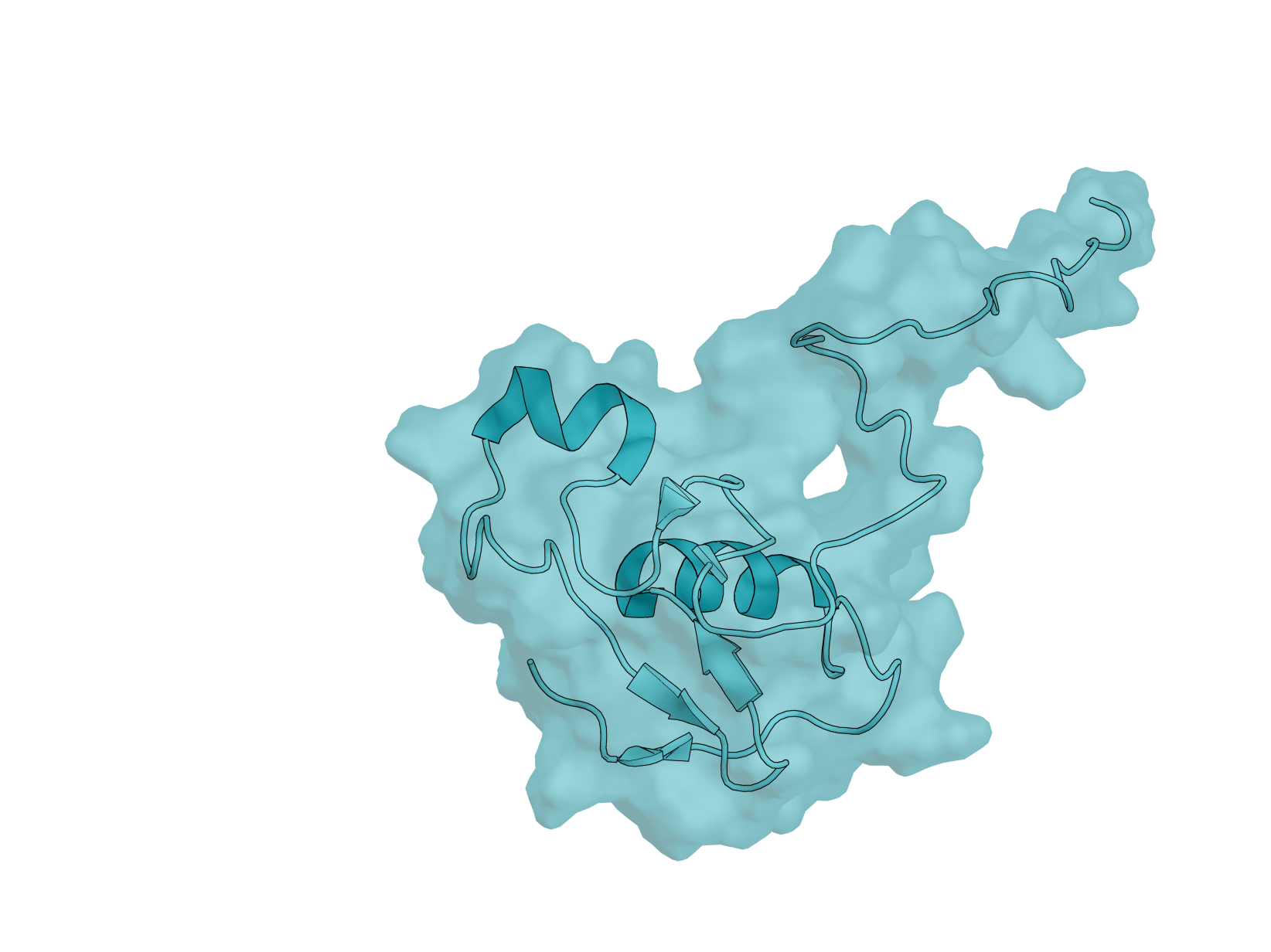
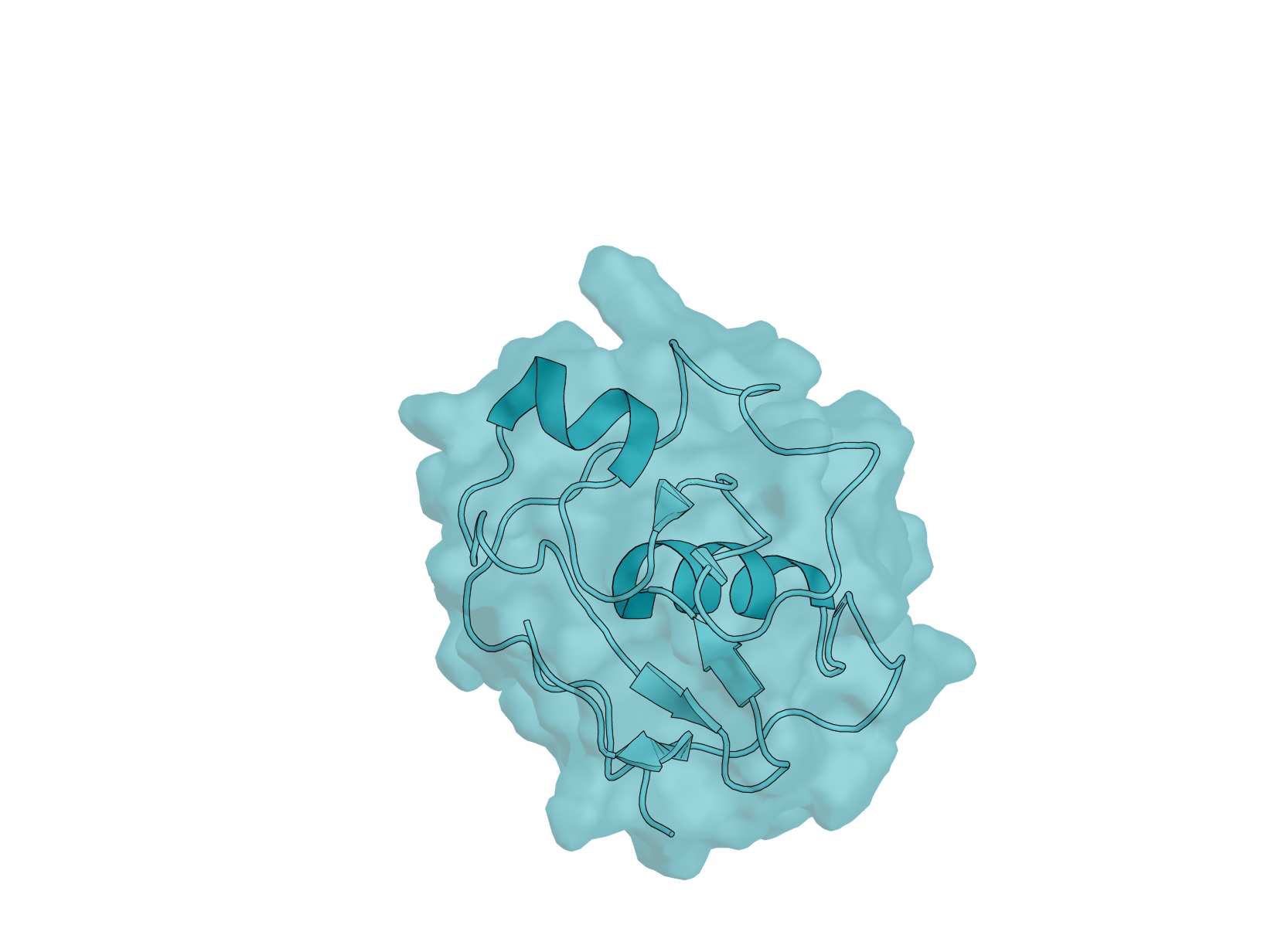
Biology: RBX-1 (RING Box Protein 1), also known as ROC1, is a 108-amino acid E3 ubiquitin ligase component essential for the SCF (SKP1-CUL1-F-box) complex. This protein plays critical roles in cell cycle regulation, signal transduction, and cancer biology—making it an important yet underexplored target for computational protein design. RBX-1 is essential for ubiquitin-mediated protein degradation, controlling the turnover of key regulatory proteins including tumor suppressors and oncogenes. It is overexpressed in multiple human cancers, and its modulation could have therapeutic applications.
Design Challenge: RBX-1 presents a uniquely challenging target for binder design. Its N-terminal region (residues 1–39) is intrinsically disordered, while the C-terminal RING-H2 finger domain is stabilized by three zinc ions in a cross-brace arrangement. This combination of flexibility and rigid metal coordination creates a dynamic surface that tests the limits of current design methods. Additionally, RBX-1 functions as a hub protein with multiple binding partners - cullins, E2 enzymes, and NEDD8 machinery - offering diverse potential epitopes but requiring careful site selection. Unlike well-benchmarked targets such as EGFR or PD-L1, RBX-1 remains largely unexplored in the binder design literature, providing a fresh proving ground for your methods.
How to participate?
Design your protein binders. Use any computational method! The designs must be de novo, which means we do not allow motif scaffolding nor lead optimization. Therefore, all binder sequences must have a minimum edit distance of 25% of total sequence length to known proteins in UniRef50.
Prepare Your Submission.
A method description (PDF, 2 pages maximum) covering background, approach, and any in-silico results.
A ranked list of binder amino acid sequences (CSV, ≤250 AA each, maximum 100 sequences per team).
Submit & Wait for Results. We will select top designs from each team by your own ranking, so prioritize carefully. Each team gets equal slots: 300 ÷ (number of teams). Adaptyv will test expression and binding affinity of these 300 designs, together with some positive and negative controls. The results will be announced at GEM workshop, and we will prepare a community-oriented write-up. We may also run follow-on functional assays for a subset of top performers and report those results as available.
Timeline
Submission Open: February 15, 2026
Submission Deadline: March 26, 2026, AoE
Results Announcement: April 26, 2026, at Rio de Janiero
Awards & Recognition
At GEM 2026, we will announce the team with the Best Designed Binder, as well as the runner-up. The teams will receive a 3D printed protein structure of their design. The top team will receive 1,000$ USD monetary award, and the runner up will receive 100$ USD monetary award.
All participants receive experimental data for all tested designs and real experimental feedback, inclusion in the open-source dataset on Proteinbase, and inclusion in our upcoming community results paper.
Helpful Readings
Lessons from Adaptyv's EGFR Competition — What worked, what didn't
Your Questions, Answered
-
-
Anyone (students, academics, industry). Individuals and teams both welcome. Every person can only participate in one team.
You can participate without coming to ICLR, but the awards will only be handed out at the conference.
-
We ask you to submit two files:
Method PDF (1-2 pages): title, team members & affiliations, contact information, brief background, brief but clear description of the method (e.g., design, filtering, ranking), and brief in-silico results. References are not included in the page limit. No other format requirements.
Ranked binder list: sequences + your ranking (and optional predicted structure files as a zip file). The CSV file should have the columns of Rank, Amino Acid Sequences, and optionally, Name/Score.
-
Standard amino acids only
Single-chain proteins, length capped (≤ 250 aa)
Novelty checks to avoid near-duplicates of published sequences. Submitted sequences must have ≤75% sequence identity to a known protein, evaluated on both the full sequence and all contiguous windows of ≥30 residues (to prevent cropped derivatives of known binders). We do not allow motif-scaffolding or lead optimization.
No tags/signal peptides, no formatting characters.
-
Yes, but describe your method sufficiently for others to understand the approach.
-
Maximum of 100 sequences per team.
-
If there are n teams, we test the top 300 / n sequences per team, based on your submitted ranking. (Example: 10 teams → top 30 per team.)
Any invalid entries (see rules for submitted sequences) will be skipped.
-
We will run standardized measurements related to binding (bio-layer interferometry) and basic expressibility signals. For details, please see https://docs.adaptyvbio.com/docs/experiment-types/expression and https://docs.adaptyvbio.com/docs/experiment-types/binding. We may additionally run functional assay data.
-
Potentially. We may write a short summary paper combining approaches + wet-lab results. If so, teams will be invited to contribute depending on scope and logistics.
-
Yes. Please acknowledge the challenge and cite the final summary if one is released
-
By participating, you agree to have your submitted sequences and their experimental results published openly on Proteinbase under the ODC-ODbL license (Open Data Commons Attribution). This means the data is free to explore, download, and use for any purpose, including commercial use and ML model training, with attribution.
Neither Adaptyv nor the GEM workshop organizers will make any intellectual property claims on your methods or design protocols. Method sharing on Proteinbase is optional. You can disclose your approach in as much detail as you like. -
For competition-related questions, please reach us at gembioworkshop@googlegroups.com. For wet-lab questions, please reach out to support@adaptyvbio.com.